Using BLAST in the command line
Blast Command Line Tutorial
We will discuss basic usage of the blast+ tools (available in Compute Canada Clusters such as Graham, Beluga and Cedar) or at the NCBI FTP).
Objectives / learning outcomes:
At the end of this tutorial you shoud be able to:
- Download databases from NCBI
- Create your own custom local database
- Do a basic/intermediate nucleotide blast search
- Change the search parameters to fulfill your needs
- Search for help regarding the available parameters
Prerequisites:
Before we start, make sure you went over your bash notes, since we will be using several of the commands we saw in prevoious tutorials. Despite I will touch briefly on the web-based blast, I am assuming you know the basics on how to make blast searches on the BLAST website.
I will discuss only the blastn (nucleotide blast) for time sake, but will comment on other types of blasts briefly.
Outline of the tutorial
- Introduction to BLAST: Types of databases, searches (this is brief, remember to brush up on this)
- Downloading databases from NCBI FTP with
update_blastdb.pl - Basic
blastnsearch - Creating local databases with
makeblastdb - Tunning
blastnparameters - Basic bash manipulation of output
Before we start
Log in into your Compute Canada account, create a folder for this tutorial (e.g Blast_tutorial), and open an interactive shell with salloc as we explained last tutorial with 2GB of memory. in the terminal type (remember to change the account for your own group if you are not in Cristescu lab):
salloc -A def-mcristes --mem=2000 --time=0-2:0:00
module load nixpkgs/16.09 gcc/5.4.0 blast+/2.6.0
update_blastdb.pl patnt --decompress
Let it run in the background, we will get back at this, but it takes some time (about 10 mins after allocation)!!
Introduction to BLAST
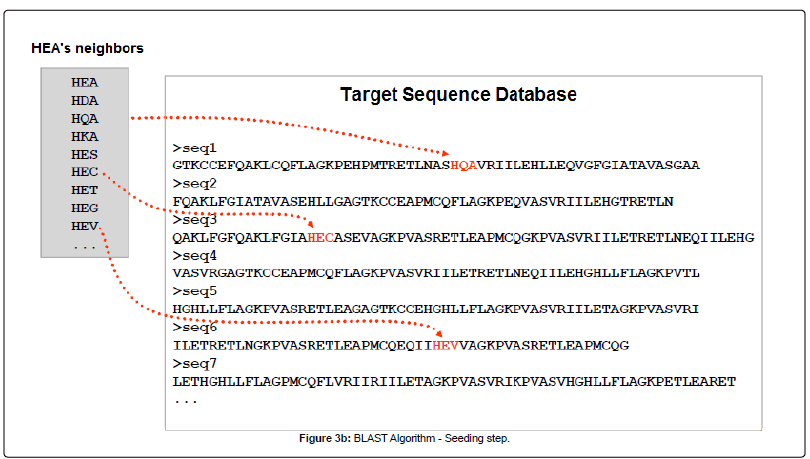
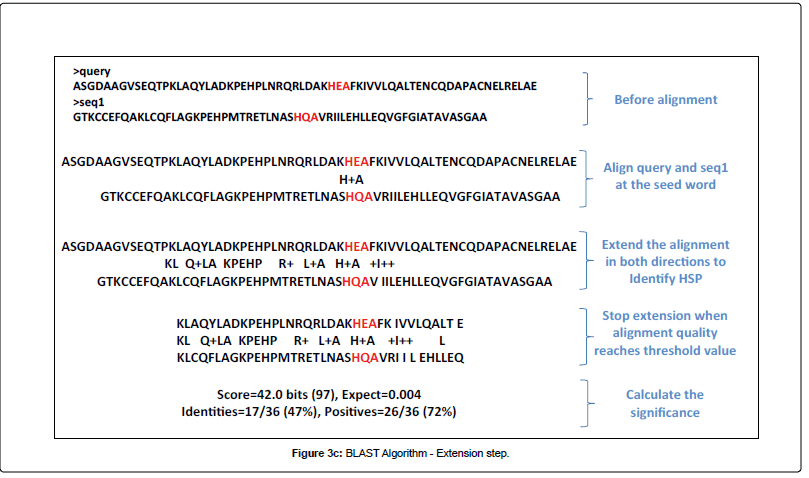
BLAST or Basic Local Alignment Search Tool, is an alignment service available at NCBI. As its name exlpains, this software aligns a query sequence (your sequence) with the database sequences, and returns the closest match to your query. It does this in 4 steps (images from Alarfaj et al.):
- Preprocessing
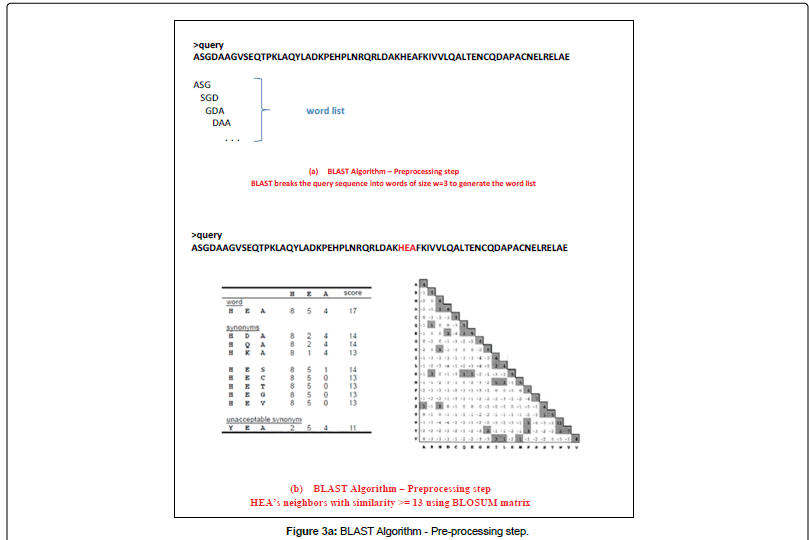
- Seeding
- Extension
- Evaluation

Let’s check their webpage and run an example blast
Downloading databases from NCBI FTP with update_blastdb.pl
Now let’s go back to the commands we typed before, by now it should be done! Let’s examine the contents of the folder, you should have multiple files with extensions .nhd, .nhi, .nhr, .nnd, .nni, .nog, .nsd, .nsi, and .nsq. You dont need to worry about their contents, but just in case:
- nhr: deflines
- nin: indices
- nsq: sequence data
- nnd: GI data
- nni: GI indices
- nsd: non-GI data
- nsi: non-GI indices
If you downloaded the sequences with the update_blastdb.pl script, you will also find a .nal file, which is an alias for the database to be searched, as well as two taxdb.* binary files with the relationships with taxonomy. However, for us to be able to search taxonomy related information we must download the taxonoy database as well. As you see we have downloaded the patnt database which is “Patent nucleotide sequences. Both patent databases
are directly from the USPTO, or from the EPO/JPO via EMBL/DDBJ”. I chose this database more for convenience, since it has more than one olume, but is small enough that we can do the tutorial with. But let’s explore what other databases we can use. For that, go to the NCBI FTP. We can see all available volumes and databases to download, but we cannot tell what they are. Let’s peak at the README file… BINGO! all the information relating these databases is sumarized in this file. Now, I mentioned that we cannot access taxonomy information without downloading the taxdb datatabase, so let’s give it a try:
update_blastdb.pl taxdb --decompress
This command should run fast since it is a smaller database. Now we have set up one database to search against. Now we only need our query file in fasta format. For easyness, I’ve uploaded a working example here. Explore this file: go to the NCBI website and find it by its accession number. Check its properties, etc. Copy the sequence and do a nucleotide blast in the NCBI website. In the Algorithm parameters tab, select Max target sequences to 500. Keep the results page open.
Now we are set to go to the basic blast.
Basic blastn search
blastn stands for nucleotide blast. It has two mandatory arguments which are -query (the fasta file) and -db the database where to look. Our first seach will be as follows (I added the optional -num_threads which makes it faster):
blastn -db /path/to/database/patnt -query /path/to/query/nameoffasta.fasta -num_threads 8 > blast.hits
Of course you have to change /path/to/database/patnt with the actual path where you downloaded the database and the /path/to/query/nameoffasta.fasta with the actual path to your file.
This command is exacly identical to:
blastn -db /path/to/database/patnt -query /path/to/query/nameoffasta.fasta -num_threads 8 -out blast.hits
Let’s check the summary of options we have:
blastn -h
Some of them are not very informative, let’s see the expanded version of the help.
blastn -help
Now we have a very verbose help, where we can see that -out allow us to redirect the output to a file. This will become handy later on when we want to manipulate files without writing all intermediate files.
The format of the output file is parwise (check the help for formatting options). You can change the formatting by passing numbers to the -outfmt option. Looking at the help we see that the options are:
0 = Pairwise,
1 = Query-anchored showing identities,
2 = Query-anchored no identities,
3 = Flat query-anchored showing identities,
4 = Flat query-anchored no identities,
5 = BLAST XML,
6 = Tabular,
7 = Tabular with comment lines,
8 = Seqalign (Text ASN.1),
9 = Seqalign (Binary ASN.1),
10 = Comma-separated values,
11 = BLAST archive (ASN.1),
12 = Seqalign (JSON),
13 = Multiple-file BLAST JSON,
14 = Multiple-file BLAST XML2,
15 = Single-file BLAST JSON,
16 = Single-file BLAST XML2,
17 = Sequence Alignment/Map (SAM),
18 = Organism Report
The default (Pairwise) shows a list of hits followed by the aligments. This is very similar to what you saw in the webpage. All query-anchored formats (codes 1-4) show the multiple alignment, in slightly different ways. The XML format (code 5) is a special hierarchichal format, similar to webpages. The tabular formats are (in my opinion) the most useful for bioinformaticians. These are tab-delimited files, which are easy to explore with head, tail, more, less and cat commands, and also are easy to parse with multiple commands and languages. We will work later on with these. Formats 8 and 9 or Seqalign format are (from the docs) “a collection of segments representing one complete alignment”, and are used for more detailed objectives, where the aligment is the result we are looking for. The format 10 or comma-separated is other very useful format since we can see it in many difefrent programs. Formats 11 - 17 are special formats that fulfill advanced usages of blast, but we arent going into their details since you probably wont use them (I havent!). The last one, Organism Report (format 18) produces an interesting table per species and all the sequence hits from that species. This can be very useful to explore taxonomy.
Now before we explore a bit more of this formats we will learn how to create our own custom database.
Creating local databases with makeblastdb
Let’s imagine hat you have a set of sequences of interest. This set can be sequenced by you or be a subset of the NCBI databases. There are many reasons why you would like to create your custom database. For example, you only want to search fish, or only prokariotes, etc.. Also, smaller database search is orders of magnitude faster that in the full database. For this exercise, go back to the web blast you made earlier. Select all seqences and download it as a fasta file. Transfer this file to the Graham. For those who dont know how to do this, we can use a command (mac and linux only) called rsync. If you are in a windows machine and you have MobaXterm, follow their instructions.
rsync -av seqdump.txt <your-username-graham>@graham.computecanada.ca:/path/where/to/move/it
**Change
Once on the cluster take a look at the file and explore it. Tell me how many lines did you find. Remember that we ask for 500? why the difference?
Now let’s see what options makeblastdn has:
makeblastdb -help
Now lets create a basic database:
makeblastdb -in seqdump.txt -dbtype nucl -parse_seqids -title Cystic_fibrosis -out cystic_fib.db
Now let’s test f our newy created tini tyni database works:
blastn -db cystic_fib -query /path/to/query/nameoffasta.fasta -out small_blast.hits
Super fast! Now we can move onto tinkering our blast searches. But before we move there, beware that we did not include taxonomic information. You’ll see what happens later. To include this, we would have to use a file that maps the accession numbers with the taxid of every entry, which is a bit out of the scope of this tutorial (if there is enough interest for this, I’ll prepare a tutorial for this).
Tunning blastn parameters
Let’s look at the blastn help again typing blast -help. We can have too many tunning parameters, here we will center in the most used ones. starting by the e-value. The E-Value “describes the number of hits one can “expect” to see by chance when searching a database of a particular size” (as per blast FAQ). This means that we want to be stringent and this number has to be small. By default is 10 so you can have e-values as high of 10. With blastn we can restrict this to any given minimum e-value:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-200
There are no hits with that restriction, let’s try a bit more relaxed (1e-200 is very stringent, even for a database as small as this one):
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-115
Ok, now we can find 34 entries with evalues smaller than 1e-115. But now lets imagine that we dont care about 30+ entries, but only the top 5. Here is where -max_target_seqs comes into play:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-115 -max_target_seqs 5
It seems that it didnt do anything. This is because all alignment formats (<= 4) don’t have this option. There is another mutually exclusive option to use in this case num_alignments:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-115 -num_alignments 5
Now we have only 5 alignemnts, although the report still contains everything < 1e-115. This is because the report have full descriptions that can be controlled with num_descriptions. Note that num_alignments and num_descriptions are not mutually exclusive, but cannot be use with -max_target_seqs:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-115 -num_alignments 5 -num_descriptions 5
You cannot see this, but it restricting he nuber of hits also makes the search faster when the database is masive.
So is -max_target_seqs useless?? Not quite, if we use the non-aligment formats this is the one we want to use. Let’s change the format for now. As I mentioned before, the most useful format (at least for me) is the tabular without comments format (-outfmt 6):
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-115 -max_target_seqs 5 -outfmt 6
Ha! there it is… now let’s check of what we have here. There are 12 columns:
- qaccver
- saccver
- pident
- length
- mismatch
- gapopen
- qstar
- qend
- sstart
- send
- evalue
- bitscore
These are the default columns, however, we can modify this however we want. Let’s imagine that we want the query name (qaccver), the hit name (saccver), the percent identity (pident), the e-value, and the query coverage:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-115 -max_target_seqs 5 -outfmt "6 qaccver saccver pident evalue qcovs"
We can even (if we have the taxdb) include taxonomy infomation (ssciname, for the scientific name of the hit):
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-115 -max_target_seqs 5 -outfmt "6 qaccver saccver pident evalue qcovs ssciname"
Lamentably, we didn’t build our database with taxonomic information. Sometimes , however, there is some taxonomic information in the subject title. We can call this one with stitle
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -evalue 1e-115 -max_target_seqs 5 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle"
Now we see some more information. Check the help in the formatting options to see all the possibilities:
blastn -help| grep -A 58 "qseqid means"
We can also filter by percent identity. To see this, let’s relax our filtering by taking off the evalue and max_target_seqs, and we will include -perc_identity as a filter:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle"
This gives us all entries with a percent identities over 90%.
**BEWARE OF THE USE OF max_target_seqs, SINCE THE OUTPUT IS NOT SORTED, THEREFORE YOU STILL MIGHT NOT HAVE THE BEST HITS. THIS EFFECT IS DESCRIBED IN Shah et al. 2018
Basic bash manipulation of output
As you could see in the help menu, not everything can be filtered directly. Here is when other programming languages and tools come into play. First let’s explore the output with the usual suspects:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" | head
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" | tail
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" | wc -l
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" | grep LT160002.1
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" | grep 'Macaca nemestrina'
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" | grep -c 'Macaca nemestrina'
Pretty useful things! :) Now, let’s imagine that you would like to collect all seqids and put them into a file. You can do this by using a nice command called cut. Basically, this command allows you to “CUT” the output based on a delimeter (by default is tab) and select the columns you need. For the example, we would like to split the file by tabs (the default) and take the second columns:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" | cut -f 2 > filewithacc.txt
I am avoiding creating a file with the output, but it will work the same way:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" -out output.blast
cut -f 2 output.blast > filewithacc.txt
Lastly (for this session), you can filter the values of columns that couldn’t be controlled directly. An example of this is the query coverage. To do this we will use an extremely flexible and useful command called awk:
blastn -db cystic_fib.db -query /path/to/query/nameoffasta.fasta -perc_identity 90 -outfmt "6 qaccver saccver pident evalue qcovs ssciname stitle" | awk ' $5 >= 90 '
This command will take the column number 5 (pident) and will print only the lines where is greater or equal than 90.
Play around with it!!
Well, that is all for now, let’s work a little bit on this and I’ll take questions you might still have!!! cheers!
EXCERCISE
In the second section of this tutorial, we would work a case study. Imagine that you have been asked to check if there are fishes in a pond. You then sampled water from the pond, filtered, extrated the DNA and amplify a 12S fragment that is of higher specificity to fish. You sequenced it using Illumina, and followed all the bioinformatic pipelines to clean it, dereplicate them, etc. You can find the resulting fasta file HERE. Using what you know of blast, can you tell if there are fishes in there? How confident are you in your results? explain HINT: use the evalue, percent identity and query coverage to guide part of your justification
For this exercise we will use the nucleotide database. If you are a member of the Cristescu lab, this will be in the Database folder at ~/projects/def-mcristes/Databases/NCBI. If you are not a member, try to use the path, if it does not work, work with somebody from the cristescu lab, to obtain the blasts you want, or download the nt databse beforehand (it takes quite a bit of time).

